Describing the Elephant - Extended H5N1 Infection Processes are Clarified Through Communications and Research - Time to Rethink an Ineffective Rationale
Dr. Greg Gray-led mdRxiv preprint study of H5N1 in 2 Infected TX Dairy Herds/Workers leads to my reflections on the timing of diagnostics to ultimately inform effective policy
We’ve all heard the story of blindfolded people doing their best to describe an elephant from the perspective of what they can feel. I think many multi-faceted epidemiological mysteries are like that in both animal and human investigations.
The most recently released H5N1 study, led by Dr. Greg Gray was published online last week: A One Health Investigation into H5N1 Avian Influenza Virus 2 Epizootics on Two Dairy Farms as a medRxiv preprint. I’d like to pull parts of the animal study data from the paper, then combine that with information gathered from other publications and sources to aggregate some of the pieces of information we have collected as “partially blindfolded” investigators and observers studying one segment or another of the entire H5N1 disease process in wildlife, spilling into cattle, then to poultry, and to a limited extent to humans. That is a long complex process, making it easy to get lost in the weeds at the many sub-steps within and between each species.
To limit the discussions this week, I want to ignore the valuable insights Dr. Gray and colleagues brought forward regarding their public health sampling efforts. Some of my thoughts regarding critical requirements for timing in sample collection apply to human sampling as well as in animals, but I’ll leave more detailed discussions to public health experts. A KFF Health News article on the Gray paper did a great job of interviewing several folks, including Dr. Gray, on the public health side regarding this paper. Please read that for more insight on the human health findings.
Back to the animal side and specifically to understanding the disease process in cattle; the published field studies all include historical and current herd clinical data in varying detail, with samples collected at the time of the farm visit - usually PCR results from milk, nasal swabs, and sometimes other samples (e.g. urine, feces).
In this study, the investigators visited 2 farms with the following reported results:
Farm A was totally recovered from herd infection at the time of the visit and sample collection; no animal samples yielded evidence of remaining viral infection, and the herd was reported to have returned to normal production. Farm B reportedly first showed clinical illness on March 20th and peaked at 14% morbidity prior to the visit on April 4th when sampling took place. At 15 days post outbreak, only one nasal swab remained positive of 20 animals tested (high CT value of 38.78) and 5 of 14 clinically ill animals had returned to negative milk sample PCR status (9 remained positive). The farm felt that its status had returned to “normal” 2 days later.
The key factor we can often overlook or minimize is the date of sampling (farm visit) versus the date of estimated initiation of clinical illness (if observed). Antigen (PCR) results are not like serology results - they can be very unstable even over 24 hours as infections progresses on dynamic surfaces like nasal mucosa. Positive nasal swab results do not remain reliably positive over long periods. Even in swine, with known proclivity for influenza virus infection of nasal mucosa, sampling guidelines suggest sampling 10 acutely ill animals to confidently determine status of a suspect infected herd.
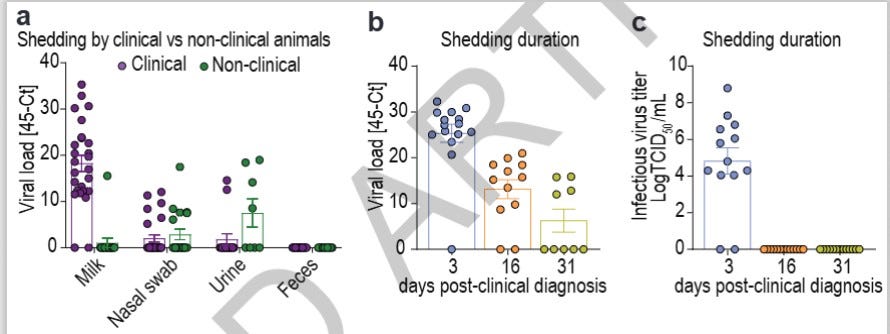
Actually, the Diehl-Dimitrov paper contains an elegant piece of work from the Ohio (#3) herd illustrating this principle better than I can explain in words:
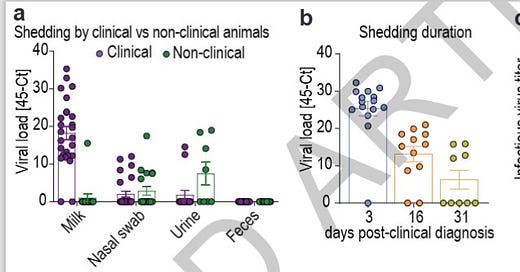
Here the researchers looked at viral loads (45-CT) from sample types and over time in the left 2 illustrations. The right graph looks at infectious virus (sample type not specified). There are some huge lessons here:
(left graph) both “clinical” (with mastitis) and “non-clinical” (without mastitis) cows showed viral loads, particularly in swabs and urine; H5N1 infection is spread widely across a herd-only a smaller percentage of the herd develops clinical mastitis. My personal extension of this information is that aerosol viral spread risks begin before mastitis develops, includes non-clinical cows, and likely extends to non-lactating dairy animals, non-dairy bovines, employees, cats, poultry, etc. early during the respiratory phase;
(middle graph) viral load drops precipitously over time (each digit on the scale is a log or 1000 times); most of the remaining positives are likely in the milk samples as the last tissues to resolve.
(right graph) infectious live virus does not last long, although this measure does not equate to transmission viability studies.
Here is additional data on paired samples (milk, nasal sabs, urine and feces) collected from animals presenting respiratory distress, drop in milk production and altered milk characteristics (clinical, n=25) and from apparently healthy animals (non-clinical, n=20) from Farm 3 were used to compare virus shedding by clinical and non-clinical animals (Extended Data Table 2).
(Note that the non-clinical animals-6/20 actually had a higher percentage of nasal shedding than the clinical animals-5/25)
Some of the very early work and pathogenic postulates regarding udder entry and transmission may have missed the significance of the early herd-wide respiratory phase of the infection, because the mastitis phase was so dramatically novel -an epidemic mastitis out of the blue tied to a novel agent. In addition, viral production in the udder is phenomenally rich, due to the receptor rich environment for influenza found there. PCR CT values in the single digits drive a lot of attention towards that tissue as the possible motherload for transmission as well as for multiplication.
When early on-farm studies and sample collections concentrate on clinically ill animals (as they should) 3 weeks after the first signs of some runny noses and red eyes (which may or may not have been noticed), we end up with red hot PCR samples for milk and a few or no weakly positive nasal swabs. Eureka, it’s not a respiratory infection in cattle - it’s a lactation-based udder infection, as shown by PCR testing!
Lots of work since then has methodically added to evidence for respiratory transmission with a subset of cows experiencing udder infections. Both the ARS heifer work and the Diehl-Dimitrov paper provide good evidence for respiratory roles. The extended time from reported first clinical illness with respiratory signs (if reported) to clinical mastitis (10-20 days) argues for a respiratory-systemic infection, followed by a mastitis sequala in up to 20 % of animals. No work I’m aware of has established reliable cow-to-cow transmission via teat orifices with commercial milking equipment.
While no one has yet demonstrated PCR in milk or mastitis in lactating cows infected intranasally, I’m not aware that such work has yet taken place. It may take several replicates, since even in the field <20% of cows in infected herds show clinical mastitis. A much larger percentage of intranasally infected cows could show positive PCR results in milk, as evidenced by anecdotal reports of positive bulk tank tests in herds up to 7-10 days prior to clinical mastitis in affected animals.
It’s disappointing to continue to see such a muted USDA response after 4 months with escalating cases and accumulating evidence of a respiratory route of infection of spillover H5 influenza (a Select Agent, for God’s sake!) into the U.S. cattle industry. I believe that the evidence shows that this is not a “special case” of a lactogenic H5 infection biologically limited to milk, udders, and bio-security lapses and cannot be eliminated with that sort of messaging or approach!
Although the infection causes relatively minor symptoms to date, except in lactating dairy cows, our U.S. emerging bio-threat decision-making process is still unable to fully fund and empower researchers and the regulatory system itself to confirm the basic scope and threat that H5N1 2.3.4.4B B3.13 poses to the cattle industry and all the other livestock industries, people, and animals that interact with it. This is not an “issue to be managed”; rather it is a huge challenge to be embraced. State and federal decision-makers need to allow our powerful research teams to utilize the tools we have available - approve and fund the damned testing and reporting!
We need widespread serology sampling to ascertain incidence across our cattle industries. We need much broader PCR and molecular-based human, animal and environmental surveillance and GISAID depositions to track viral mutations. Once we understand the scope of the issue, the rate and threat of spread, and our points of vulnerability, we can begin to develop appropriate responses.
Most experts who want to see the whole elephant have removed their blindfolds, compared notes, and are ready to move forward. It’s time for the remaining decision-makers thinking that they can still manage this nasty virus through “lactating dairy cattle messaging” to join the rest of community. The only thing being exhausted right now is the status quo.
John