FMD in Germany News Flash-Then Back to H5N1 B3.13 and a Shifting PB-2 Segment
Anxious world awaits more epidemiological details from Berlin area
We awoke Friday morning with an unnerving announcement from the Frederick Loeffler Intitute in Germany: FLI confirms foot-and-mouth disease in Brandenburg water buffalo | Friedrich-Loeffler-Institute On Saturday Loeffler announced the virus serotype: FMD outbreak in Brandenburg: serotype O detected | Friedrich-Loeffler-Institute
On Monday morning the following statement was posted by The European Federation for Animal Health and Sanitary Security (FEFASS) on LinkedIn: Foot and Mouth Disease in Germany| LinkedIn
Meanwhile, Sunday morning I came across a translated dispatch from Germany that provided several more details:
Pigs to be culled near cases of foot-and-mouth disease in Germany:
After the highly contagious foot-and-mouth disease (FMD) was detected in a herd of water buffaloes in the eastern German state of Brandenburg, pigs in a nearby region are also to be culled, officials said Friday.
The deputy district administrator of Märkisch-Oderland, Friedemann Hanke, said that all cloven-hoofed animals within 1 kilometre of the pasture with the affected water buffalo herd would be killed.
This includes a pig farm with around 200 animals in the neighbouring district of Barnim, as well as four sheep.
The outbreak in Märkisch-Oderland, not far from Berlin, represents the first known cases of FMD in Germany since 1988, and the first ever case in the state of Brandenburg. The country and the broader European Union have been considered free of FMD in recent years.
Three water buffaloes that died in the town of Hönow were confirmed to be infected with the disease, Brandenburg's state Agriculture Minister Hanka Mittelstädt announced on Friday morning.
The remaining 11 water buffaloes in the herd will be killed and disposed of as a precaution to reduce the risk of further spread, according to local officials.
Within a restricted zone with a radius of three kilometres, samples will be taken from all animal husbandry facilities with even-toed ungluates - animals with an even number of hooved toes - said Deputy District Administrator Hanke.
In a wider surveillance zone, there will be random sampling. The transport of animals and feed is prohibited in these areas, according to Hanke. In addition, information signs are to be set up in the zones.
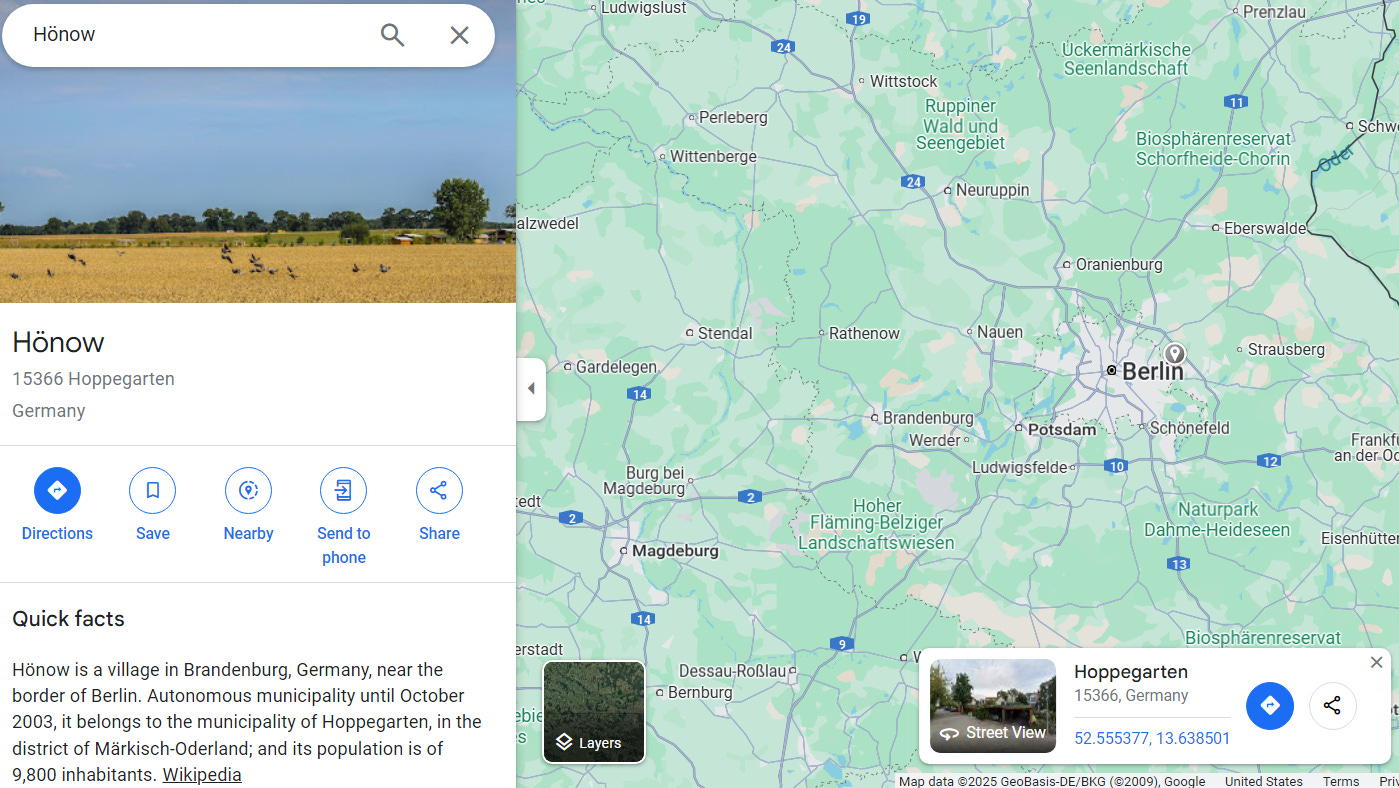
Here is a map of the Berlin area with the village of Honow marked with a small gray marker:
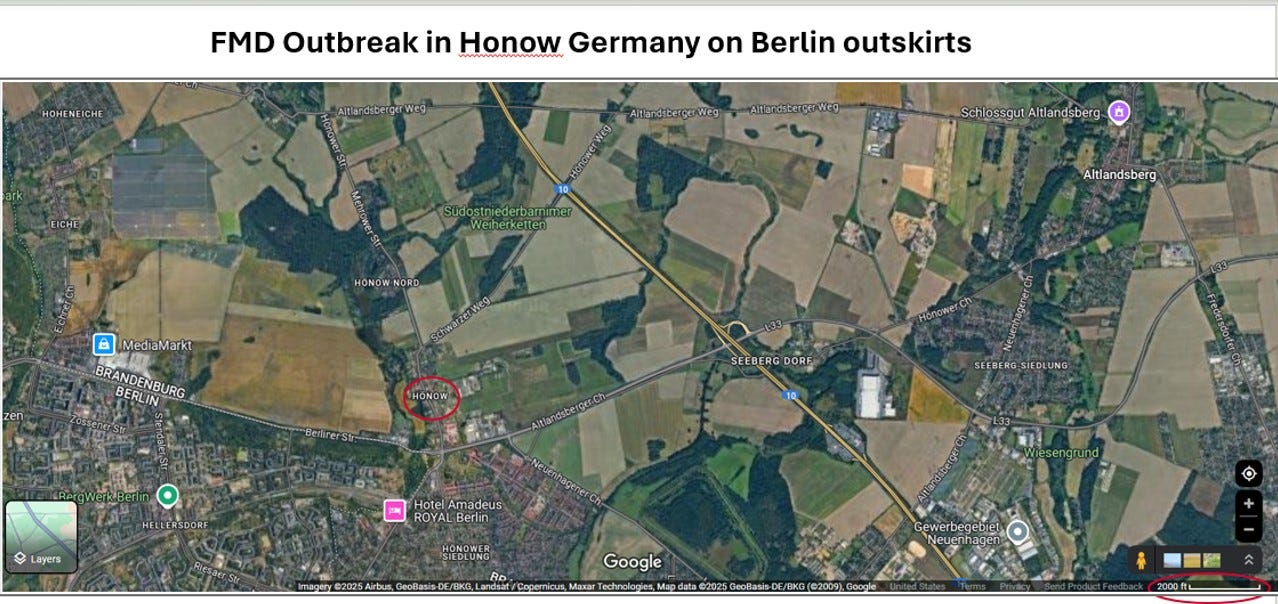
Here is an aerial map showing the topography of the area. The village rapidly gives way to pastures and fields to the west, north, and east. I circled the scale on the bottom right corner - 2000 feet.
The affected herd of 14 pastured water buffalo suffered 3 mortalities and is located less than 1 kilometer (.6 mile) of 200 hogs and 4 sheep which are also now depopulated. The pig farm is in Barnim district (versus Märkisch-Oderland district), for those keen on really narrowing down possible locations…
We’ll learn much more this week, but 3 things concern me:
discovery of the case through dead animals - How many days was the herd of 14 water buffalo incubating infection before 3 succumbed to infection?
proximity of 200 swine - Pigs are notorious FMD amplifiers and shedders. If that herd was infected and shedding virus clinically or sub-clinically, spread could be wider.
winter climate and aerosol spread. Here is a paper discussing potential for spread of FMD via airborne transmission: Airborne Transmission of Foot-and-Mouth Disease Virus: A Review of Past and Present Perspectives - PMC
I really have no idea regarding the agricultural structure in Eastern Germany, although the map appears to show widespread hay and/or pasture or crop areas. Potential for spread from the initial herd or herds will depend upon: 1) animal density in the immediate area; and 2) customary marketing and trading practices for local live animals and products. Germany (and neighboring Poland) officials are already struggling mightily with periodic ASF outbreaks from wild boar and boar meat incursions into domestic swine herds. Adding FMD to their plates is another burden they surely don’t need at this juncture. Hopefully this will remain local and contained, but get the vaccine plans ready…
Ongoing H5N1 B3.13 Polymerase Mutations
I want to devote the balance of this blog to a Michael Coston post I initially overlooked: Avian Flu Diary: Preprint: Polymerase Mutations Underlie Adaptation of H5N1 Influenza Virus to Dairy Cattle And Other Mammals. I reread it this weekend and more fully considered the true impact of the research findings. Here is the paper in full:
In the introduction the authors’ state:
…Avian-origin influenza viruses require polymerase mutations to utilise the shorter mammalian ANP32A/B proteins23. The most widespread, and best characterised adaptive mutations are those in the PB2 subunit of the polymerase including PB2 E627K22, Q591R/K24,25, and D701N25,26; all of which have been detected during other mammalian H5N1 outbreaks5–11 but are absent in USA dairy cattle viruses. However, alternative adaptive mutations in the influenza polymerase that enable the virus to co-opt suboptimal ANP32 proteins are possible27–29. Here, we investigated potential mammalian adaptations in the polymerase of cattle-adapted H5N1 viruses and identify two key mutations that allow the B3.13 virus to replicate in cells from cattle and other mammalian species, including humans.
The initial issue for researchers has been that the B3.13 “dairy” strain has lacked traditional PB2 subunit mutations to allow mammalian adaptation. So they set out through phylogenetic analyses to identify alternative common mutations among sequenced isolates to explain successful adaptation.
Abstract
In early 2024, an unprecedented outbreak of H5N1 high pathogenicity avian influenza was detected in dairy cattle in the USA. The epidemic remains uncontrolled, with spillbacks into poultry, wild birds and other mammals including humans. Here, we present molecular and virological evidence that the cattle B3.13 genotype H5N1 viruses rapidly accumulated adaptations in polymerase genes that enabled better replication in bovine cells, as well as cells of other mammalian species including humans and pigs. We find evidence of several mammalian adaptations gained early in the evolution of these viruses in cattle including PB2 M631L, which is found in all cattle sequences, and PA K497R, which is found in the majority. Structurally, PB2 M631L maps to the polymerase-ANP32 interface, an essential host factor for viral genome replication. We show this mutation adapts the virus to co-opt bovine ANP32 proteins and thereby enhances virus replication in bovine and primary human airway cells. Importantly, we show that ongoing evolution during 2024 in the PB2 gene, including a convergently arising D740N substitution, further increases polymerase activity in a range of mammalian cells. Thus, the continued circulation of H5N1 in dairy cattle allows virus adaption improving replicative ability in cattle and increasing zoonotic spillover risk.
I have made the mistaken assumption based on “no news” and relatively homogenous phylogenetic trees that no significant changes are occurring in B3.13 virus as infections proceed over time. However, this paper shows that “ongoing evolution during 2024 in the PB2 gene, including a convergently arising D740N substitution, further increases polymerase activity in a range of mammalian cells”. A convergent substitution can be defined as a random mutational change that provides an organism a competitive advantage. Thus, as the same mutation “succeeds” in multiple infections, it converges as a common change in the phylogeny of the virus in multiple sequences.
What really caught my eye in rereading this paper was the conclusion of the Discussion section of the paper:
What is the current risk to humans posed by the cattle virus? Our results show that the cattle virus containing mammalian adaptations can replicate better in mammalian cells than the minimal avian-like precursor virus and is therefore a clear risk to mammals which have consumed infected milk such as cats and raccoons13. Thus far, these cattle viruses show inefficient airborne transmissibility in ferret experiments30,44 and have not adapted to transmit via respiratory route or to efficiently use human receptors (α-2,6-linked sialic acids)45–48. Nonetheless, frequent exposure of an antigenically novel, highly pathogenic influenza virus that can replicate well in human cells is concerning. Our data suggest that the emerged cattle influenza virus is also capable of efficiently infecting avian and swine cells, further increasing the risk of spill over from cattle to other species. In fact, the mammalian-adapted virus appeared to have no fitness cost in avian cells, explaining the high propensity of this virus to spillover into poultry, and suggesting such a virus could maintain its mammalian adaptations if it spilt back into wild birds. With each human infection and increased polymerase activity leading to higher levels of replication, there is a danger of further evolution changing viral receptor properties. Additionally, a reassortment event with human seasonal influenza viruses could lead to a novel virus, particularly during the Northern hemisphere winter influenza season.
Recent experimental infections in cows suggest that the ability of the North American B3.13 cattle influenza virus to spread via milk is not unique and that other mammalian viruses are capable of transmitting via this route16. Phylogenetic data suggests that this originated from a single spillover event from wild birds to cattle and such spillovers must be rare as IAV has rarely been recorded in cattle. Nonetheless, in the absence of an effective control strategy, the high pathogenicity H5N1 virus may now become endemic in US dairy cattle, requiring continuous monitoring even in the absence of overt disease. Moreover, many other clades and strains of H5N1 virus continue to emerge through reassortment, causing zoonotic infections. For example, the emergence of the D1 genotype of H5N1 virus in North America has caused a recent uptick in spillover from birds to mammals including humans and swine49. Urgent development and testing of broadly reactive H5 influenza vaccines for both animals and humans is a priority.
H5N1 B3.13 and D1.X have proven their transmissibility and genomic flexibility. The big California dairy epidemic headlines may be ebbing, but this virus is still around. Michigan had a positive bulk tank again in late December, and we’ll likely find some more as National testing ramps up. Time will tell whether partial herd immunity prevents large acute herd outbreaks; however, it’s a long way from a “quiet herd” to a fully negative herd with influenza virus in an extremely movement-dependent industry.
If you want to quiet a room of dairy producers (or pork or beef producers), ask them how they would manage business continuity if the “bovine influenza” in Texas last February had been FMD. All those comments about the “impossibility” of stop movements and movement testing still hold for every industry. We essentially defined away H5N1 as an actionable agent for cattle because we couldn’t manage the consequences. So, what’s the plan for FMD? What if Honow Germany is next door?
John
Scientists Are Racing to Develop a New Bird Flu Vaccine | TIME
mRNA Vaccine Provides Broad Protection against All Known Influenza Subtypes
No need to provide an avian flu vaccine for humans yet, expert says • Louisiana Illuminator
2 people sick, 9 monitored after H5N1 bird flu hits Oakland Co. flock