H5N1 2.3.4.4b D1.3 in Ohio-Indiana Poultry Outbreak with Associated Undisclosed Human Sequence Data
Background phylogenetic work on USDA raw sequence submissions reveals a surprise unpublicized emerging avian genotype (D1.3) as a common source in the large multi-state poultry outbreak
I start every week wondering what will develop in the H5N1 saga that could possibly be of more interest than already published information. This week I had a lengthy and fruitful conversation with an extremely well-informed reporter, Nat Lash, with ProPublica. Following the conversation, he sent me an e-mail with the following information:
Hey -- thanks again for being down to chat. Dug a little deeper and a minor correction to what I suggested on the call, which I find rather interesting: the mercer/jay/darke county outbreak is actually D1.3, so it didn't come up on the Nextstrain build of D1.1 but are still assembled by Anderson Lab into the consensus sequences.
Upon receiving this information I contacted a couple of esteemed molecular biologists I’ll leave unnamed who provided me the following information:
The “Andersen Lab” refers to my friend Kristian Andersen at Scripps who has provided a very helpful resource on his github page that creates consensus sequences from the SRA data USDA submits (which are raw reads that are very messy for analysis).
https://github.com/andersen-lab/avian-influenza
The tree from Nat Lash in your email appears to be a single introduction into poultry because there are no wild bird sequences in that section of the tree, but I don’t know what Nat used as his background data set and whether it included all wild bird samples that are available, which makes a huge difference for interpretation. I heard all the Western Ohio poultry cases were D1.3. Really new genotype, not much known about it.
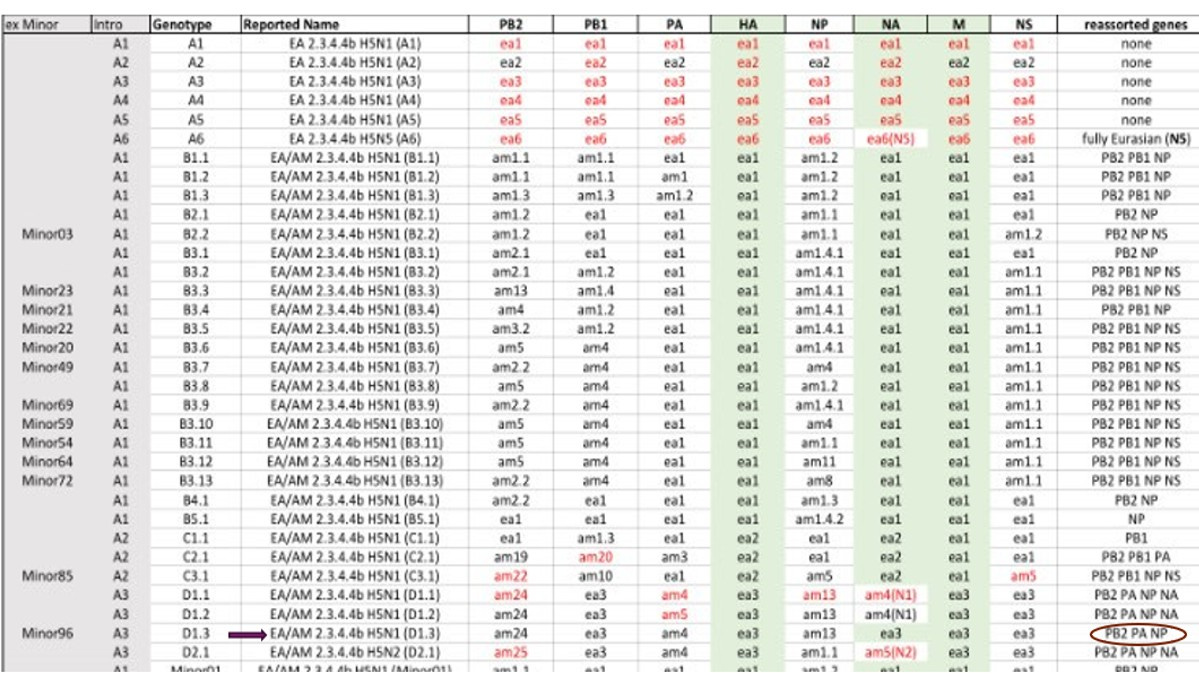
I also received access the following table which shows gene segment reassortment origin changes for each of the 8 influenza segments for the various H5N1 genotypes. None of this is meaningful to me, expect that it is illustrative of how the Genotype nomenclature system is derived. If I read this correctly, the D1.3 genotype has swapped out a Eurasian N1 for a North American N1 found in D1.1 genotypes. The red versus black colors on the coding is also likely significant?
Nat Lash had also done searches for D1.3 and stated: “It does appear D1.3 has been around since at least Nov, popping up in MN, PR, and on another branch with no state data in ducks & geese. APHIS reports only one detection in wild birds in this area around then, a Canada goose mortality sampled by OH DNR on 12/14/2024, but not seeing anything in my build that's an obvious match.
In summary, H5N1 2.3.4.4b D1.3 is a new genotype, found sporadically, including perhaps once in Ohio in December, but not matching the sequencing on the outbreak strains isolated. While the outbreak may well have originated from a wild bird spillover into a poultry flock, no one has deposited sequences to date that would indicate that path, or intermediate wild bird pathways for spread between the poultry flock outbreaks.
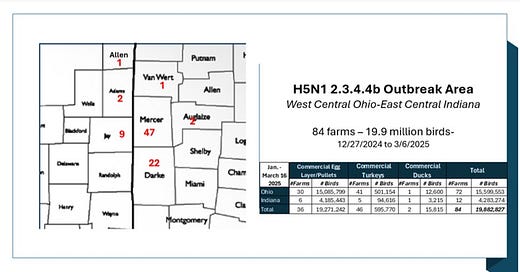
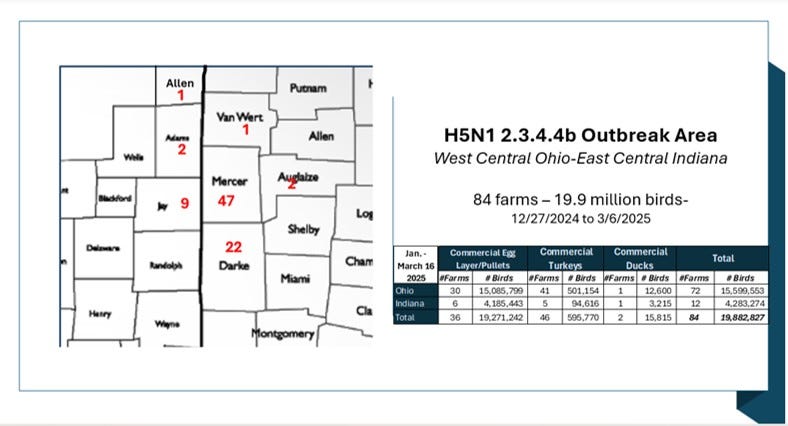
The outbreak may not be completely over; however, no new cases from the affected area epicenter have been reported since March 6th. Here is the county level case count as available from state animal health web sites:
It’s been a staggering loss in a very short period of time, hitting an extremely high percentage of poultry flocks in the area. I calculated that Ohio’s Mercer and Darke Counties alone lost over 98% of layer hens listed in their NASS 2022 inventories! Even if their inventory has grown somewhat since then, that is still a huge attack rate in a small area on an industry that in general provides fairly robust biosecurity measures against wild bird entry into larger laying facilities. (Hint - think likely aerosol/area spread versus biosecurity breaches with wild birds- see my earlier columns.)
We don’t have access to temporal and geospatial data matched with outbreak sequences. Nor do we necessarily have the latest sequencing results for all wildlife samples collected in the area. Regardless, it is troubling to have such a widespread poultry outbreak in such a concentrated area with no wild bird samples to anchor the phylogenetics at this point. The picture will undoubtedly resolve somewhat over time with more sampling and analysis. Hopefully both animal health and wildlife agencies will quickly release D1.3 genotype sequences and accompanying metadata as widely and rapidly as possible to fill out this picture.
Another outstanding question relates to mammalian adaptation for D1.3. We assume that the one reported hospitalized human H5N1 patient, a poultry worker, was likely infected with this genotype. It’s extremely troubling that CDC has reportedly sequenced this virus, but to date has failed to publicly disclose its findings or share the sequence(s) to GISAID.
I’d also assume that APHIS Wildlife Services or other agencies are actively sampling peridomestic species (mice, rats, cats, etc.) in close proximity to the outbreak flocks. Is this virus, widely dispersed across 84 farms in 2 months spreading to other mammals? Is D1.3 carrying back into wild / migrating bird populations to put other poultry populations at risk?
Finally, what is the status of the NMTS in Ohio and Indiana? Have samples been drawn, tested, and reported to officials from milk silos serving dairy herds in the areas with widespread poultry outbreaks? Have individual dairy herds, beef herds, or swine herds been sampled, especially if showing any signs of illness? Positive answers to these questions paired with negative results would go far in lowering the concerns of many.
I don’t want to drone on. We have a relatively new zoonotic H5N1 2.3.4.4b D1.3 genotype that has killed over 19 million chickens on 84 farms in a small area in less than 2 1/2 months! It put one poultry worker in the hospital. No one has even bothered to publicly announce the new genotype to my knowledge! CDC has likely sequenced it from the hospitalized human patient yet has failed to deposit the sequence or even inform the public of its findings.
Yes, I’m pissed! Has the One Health world gone mad? Am I missing something??
John





I don’t believe the USDA has released any figures to date on how many cows have died from H5N1, nor numerous other important details with regard to how the virus is affecting herds in the various states. Correct me, please, if that’s mistaken.