H5 Wastewater Sampling - What is it Good For?
The early bird catches the worm - we can "see" incubating outbreaks before they are apparent - can we bend our human "protocols" to accommodate that power?
I’ve been a fan of environmental sampling for early detection of infectious diseases since I became aware of its utility for early detection of new outbreaks of COVID-19 ahead of other more traditional clinically based detection methods. Large volumes of viral shedding precede clinical illness for many of the most infectious viral disease in both people and animals and certainly precede the lag from clinical awareness to the time of successful diagnostic testing and reporting that is inevitable under the best of circumstances. The molecular diagnostics revolution in both agent amplification and purification has made it possible to detect agent presence in very small, contaminated pooled samples. Unfortunately, our willingness to actually explore and utilize the technology to its full potential has not kept up with its capabilities yet, especially in animal agriculture.
This was somewhat embarrassingly illustrated us to by a research preprint released earlier this year demonstrating H5 RNA in wastewater samples from Texas cities with milk processing facilities: Detection of hemagglutinin H5 influenza A virus sequence in municipal wastewater solids at wastewater treatment plants with increases in influenza A in spring, 2024 | medRxiv
This work shows that wastewater monitoring can provide an early warning for outbreaks likely to produce contributions to the sewer shed outside of the expected human-associated inputs, including for animal outbreaks of diseases with zoonotic potential. In Potter/Randall County, retrospective testing shows that H5 was detectable in wastewater on Mar 1, 2024 - nearly a week before an unspecified disease was reported in dairy cattle in Texas (Mar 7) and several weeks before the causative agent was identified as H5N1 (Mar 25).
These results in part led CDC and public health partners to expand H5 monitoring efforts across the U.S. as the H5N1 dairy influenza crisis spread across multiple states, and subsequent studies showed 20+% of retail milk samples were positive for (killed) H5 RNA. The whole molecular testing process in “pooled” agricultural samples forced the dairy industry and its regulators to admit that H5 infections in the national dairy herd was (and is) much more widespread than our limited traditional outbreak testing and tracing processes can manage in today’s complex industry management and animal movement structures.
Initially H5-specific RNA assays for wastewater samples were not widely validated and available; however, non-H-specific influenza matrix gene assays were available for assessing generalized influenza levels. These assays served well as substitutes for H5 assays in late spring through the summer because seasonal flu declines to near 0 in the human population during this period. CDC monitored Matrix levels in a series of wastewater plants, and several did show elevated influenza readings, mostly in areas with known dairy infections, but also in some states/areas without known dairy herd infections, leading to questions regarding whether those samples might contain H5 of dairy cattle origin.
This is where follow-up execution seemed to lag. While CDC dutifully reported the elevated influenza readings on their web site, state and local public health authorities seemed unwilling and unable to actually characterize the samples to determine genotype(s), speculating on possible avian origins without further characterizations of the elevated influenza assays. Animal health authorities ignored the results, since they were not official tests on animals covered under livestock regulations. That state of affairs continues currently, even after wastewater testing has evolved to H5-specific assays. We continue to hear speculation about possible wild bird contamination of sewage sources, but no willingness to actually genotype the sewage samples to separate out dairy clades from likely wild bird sources of positive samples.
Assuming a sample is shown to be H5-positive, the other critical part of the analysis relates to analysis of the sewage flow into the plants where samples are obtained, somewhat analogous to a disease “traceback” investigation. In the current outbreak with widespread dairy cattle infections, milk remains a primary suspect for causing high H5 levels (even in “negative states”, because cows shed such high levels of virus in milk for relatively extended periods and widespread bulk tank testing is not yet mandatory. That is why it is so important to determine the H5 genotype of positive wastewater samples first if possible, to determine if milk is the likely source as evidenced by B3.13 signatures in the genotype.
Then if confirmed as B3.13, it is important to realize that the sewage plant is likely tied to milk or milk product processing, distribution or disposal, not geographically to dairy herds themselves! The infected cows and herds are likely elsewhere, and herd locations can usually be estimated to some degree with processor records and/or public knowledge of likely milk sheds for processing plants. The degree of geographic specificity collected/shared will be determined by state animal health officials and legal constraints, but public health officials should at least have some idea of where risks for zoonotic H5 infections in dairy workers may exist to allow for enhanced monitoring for human clinical illness in higher risk areas.
I have already pointed out a possible signal from Harrisonville, Arkansas, in a blog on September 14th: Human H5N1 Case(s?) Information from Missouri Continues to Unfold Late Friday. This sample may have had epidemiological ties to the human case in Missouri which lacked any documented animal contact. The evidence tying this wastewater sample to that human case is far from conclusive! However, it would have been useful to compare sequences between the partial genomic data from the human case obtained by CDC and whatever could have been obtained from the Arkansas wastewater sample, which was possibly tied to southern Missouri-eastern Kansas milk supplies distributed through an Arkansas facility in Harrisonville.
Contemporary Examples in Wastewater Scan data:
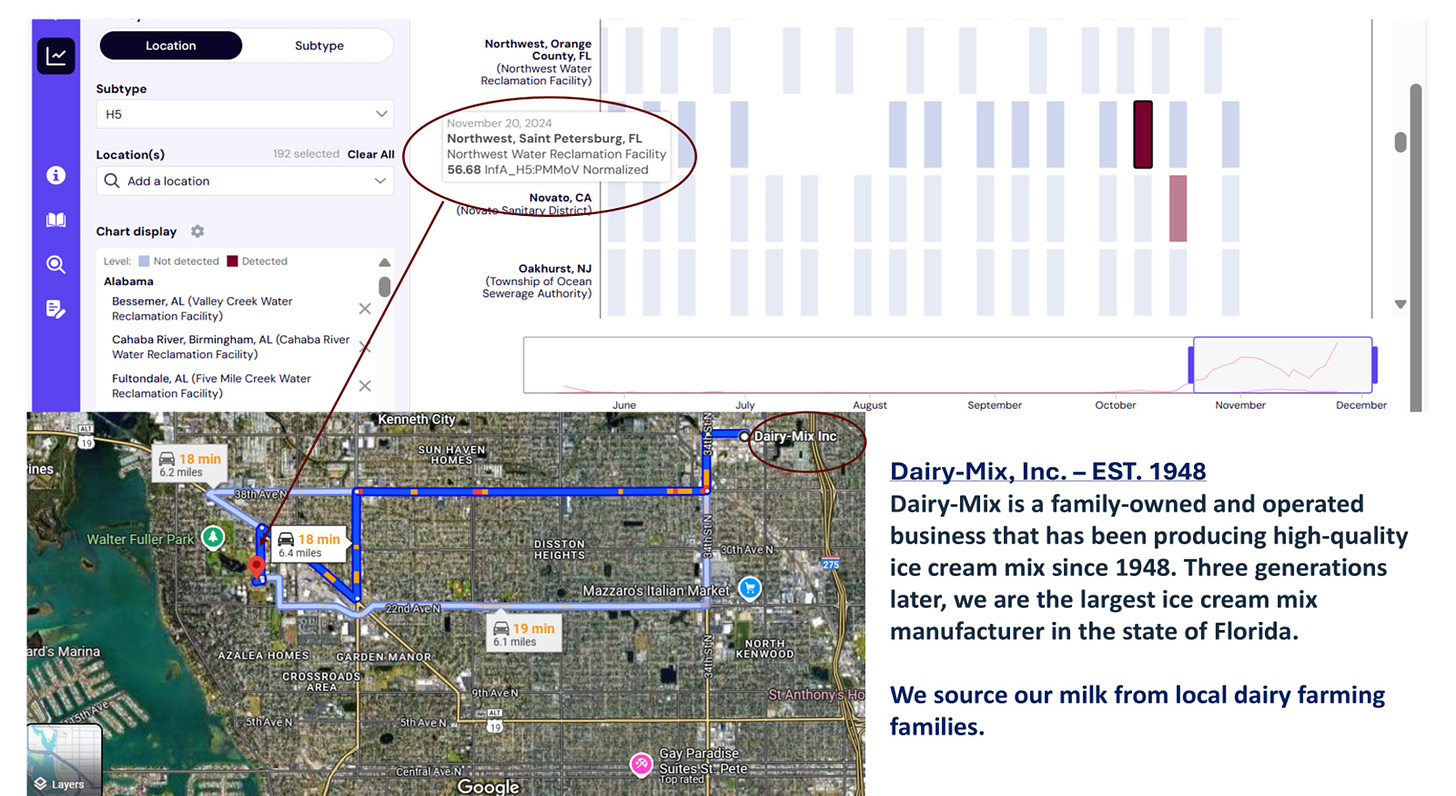
Here is an “outlier” in the current Wastewater Scan Dashboard that shows a positive reading from November 20th in St. Petersburg, Florida. This sewage plant is located in St. Petersburg about 6 miles from the largest ice cream mix plant manufacturer in the state, which advertises receiving milk from local dairy farming families on its public website.
Florida has NOT reported any clinical H5N1 infections in dairy herds to date. It’s possible that this reading is not due to H5N1 B3.13, but has anyone tested? It’s also possible that wild birds may nest somewhere in the sewage area - if so, the positive sample won’t genotype as B3.13. It’s also possible that this ice cream plant doesn’t drain into this sewage plant - a quick check of city records will rule that plant in or out as a possible source of waste milk into the sewage stream of the plant.
All of these basic steps should be routine work as part of the H5 wastewater monitoring system. Every “hit” should have genotyping attempted as a condition for participation in the national program. Further epidemiological investigations may or may not be warranted pending genotype results and wastewater sewage flow analysis. Given past policies with COVID, I’d be surprised if Florida chooses to act on this signal, but wastewater data collected with federal funding should at least be followed though to sequencing attempts on H5 positive samples with deposition of successful genotypes into GISAID or GenBank, as appropriate.
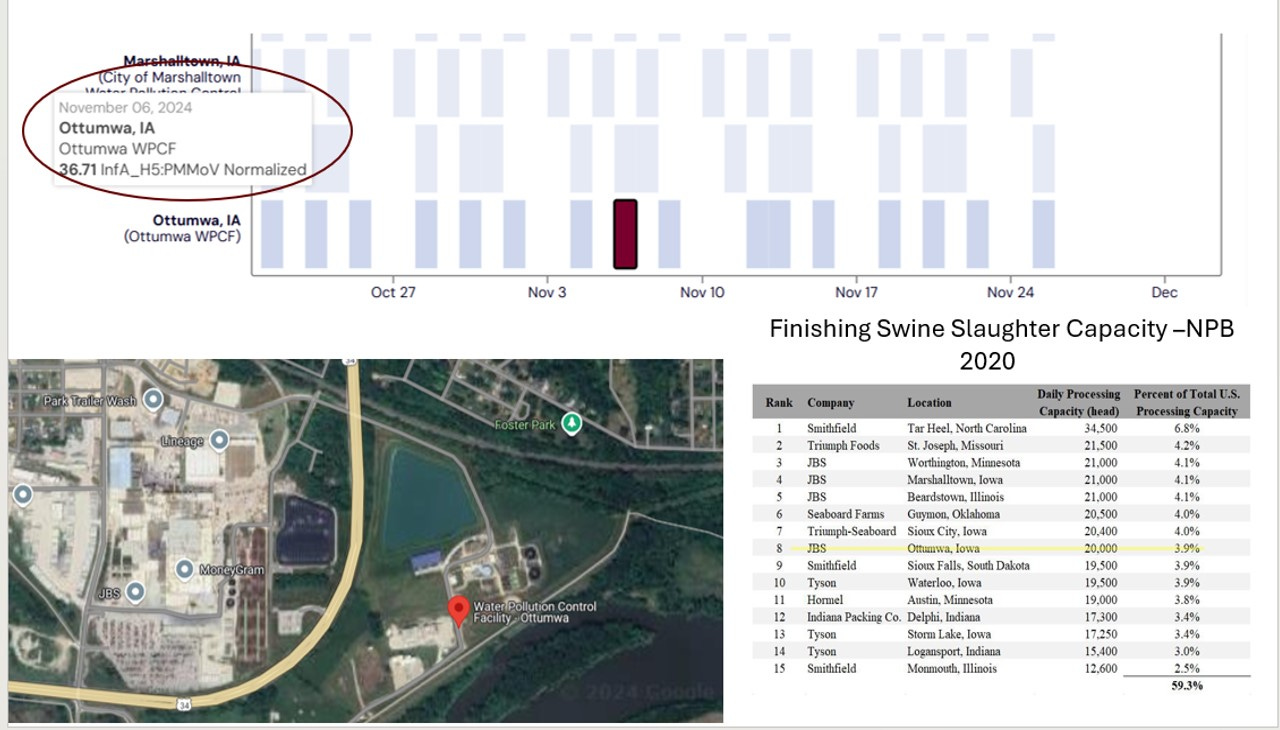
Here is another “hit” from Iowa in early November with another entirely different possible implication, depending on the genotype of the sample:
Back on November 6th, Ottumwa, Iowa recorded an H5 reading of 36.71 H5 units per dried gram of solids. The sewage plant is adjacent to a “top 10” JBS finishing swine slaughter plant. I don’t know much about this plant related to sewage flow and wild bird “contamination” risk at this sewage treatment site. However, given the swine slaughter plant, I’d hope that genotyping of the sample would be of high priority to investigate any risk of H5 viral RNA in swine slaughtered at the plant. If the sample is 100% avian in origin, it’s likely an avian-based contaminant in the sewer-shed. However, if there should be some mammalian adaptations or reassortant segments in the sample sequencing, we have a much more serious possible swine cross-over virus genome to further investigate. The point is - sequence the finding to determine the risk!
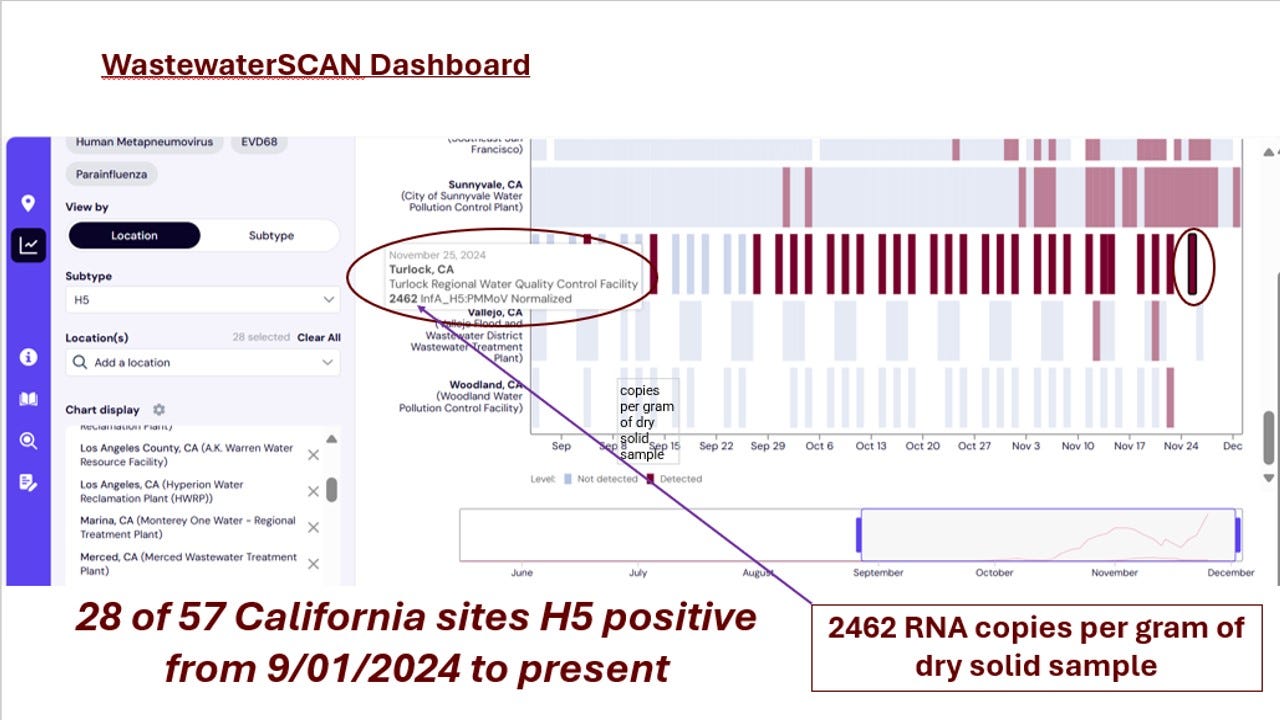
Finally, let’s consider the balance of the positive findings (all from California) on the current Wastewater Scan Dashboard. I can’t show them all in one screen shot, but here is the link if you want to see it yourself: WastewaterSCAN Dashboard-California.
28 of 57 sites have been positive since September 1st! I highlighted one sample in Turlock below, where a lot of milk is processed, with the highest reading I have seen - 2462 copies of RNA per gram of dried solids:
I’ve seen a lot of uninformed speculation online regarding what all these positive readings mean regarding human infections in California. My first reaction is…not much. We are in the midst of a huge outbreak in dairy cattle and poultry in the state. Mice and other peridomestic species are also undoubtedly highly infected. Until epidemiologists first sequence the samples, then analyze every sewage site by genotype(s), possible milk sources, poultry sources, and wild bird sources from any open water inflow into sewage treatment, the gross H5 readings are impossible to decipher by themselves. Human incidence analysis may require diagnosis by subtraction of animal sources from the total results. The tool will not be truly valuable for public health until we all have a good way to account for background H5 levels in the stream due to non-human contributions. That is my final point - H5 wastewater surveillance for H5 is a true One Health tool that is not useful for either public health or animal health stakeholders (including wildlife) without full participation and collaboration from experts in all areas.
Finally, animal health stakeholders need to embrace this technology as a valid early detection methodology in production agriculture. Environmental sampling techniques allow diagnosis before anyone suspects there may be a problem. That’s a hard sell for a regulatory system wedded to decision-making based on molecular-based confirmatory diagnostics in individual animals already showing case-compatible clinical signs. Environmental sampling removes diagnostic power from the regulatory veterinarian / centralized lab structure and places it in the “sewer”. While disease outbreaks and spread can be documented much earlier, our disease control “managers” lose control of the narrative. We shouldn’t underestimate the resistance to this loss of diagnostic process control caused by an otherwise common-sense early detection screening method. Our animal agricultural industry is driven by export markets dependent upon orderly management of disease status communications and doesn’t respond well to unexplained “surprise” disease findings in sewage or other environmental samples.
The potential payoff is that the benefits of much earlier detection far outweigh the challenges of dealing with the surprise and initial uncertainty regarding the sensitivity, specificity, actual location, and extent of infection represented by a positive environmental sample. Infections are not confirmed until a formal case definition is met in an animal at a single location, including positive confirmatory testing at an approved laboratory. We should use the environmental sampling to leverage initial case finding and outbreak assessment, not to replace the initial confirmatory process. If we can get past the trust and communications expectations issues with placing environmental screening into a much broader range of sampling situations, such as wastewater, our chances for economically detecting devastating high consequence diseases in their early stages will increase greatly.
We may be more “out of control” in the short-term with positive environmental sampling, struggling to define infection sources and formally confirm a diagnosis, but the trade-off is a smaller outbreak, diagnosed as it first unfolds and before it spreads in our modern multi-state production systems. As the H5 dairy outbreak has shown, we will inevitably move to environmental (mass bulk tank sampling) at some point anyway to deal with large outbreaks. Why not start before the outbreak occurs, saving much larger economic and human losses from a delayed diagnosis?
John